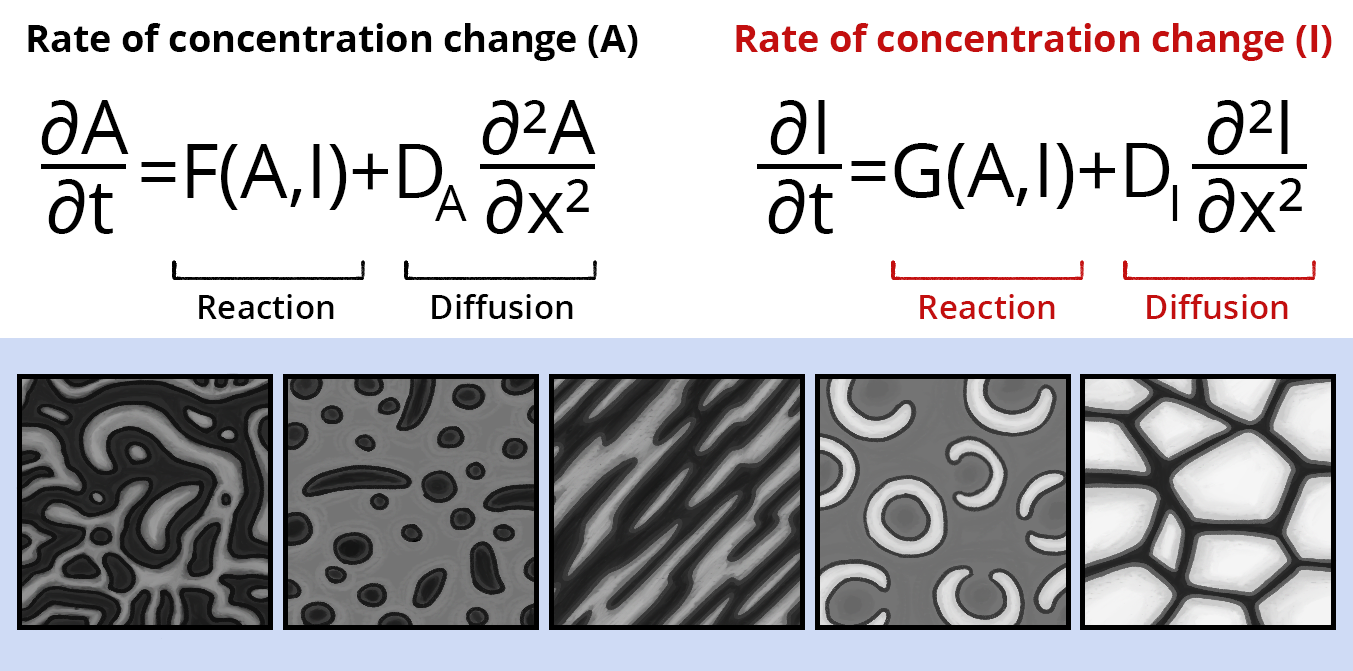
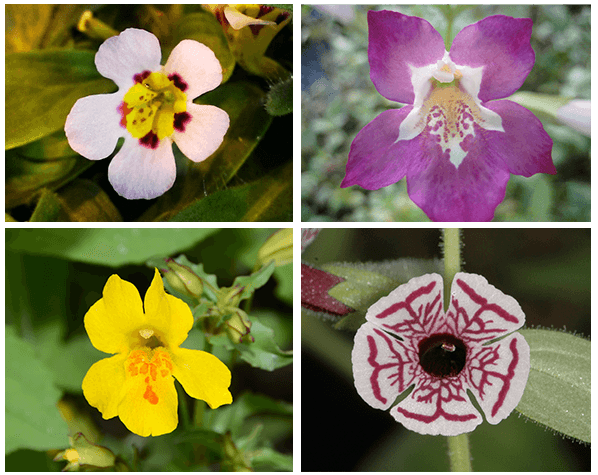
References
Ding, B., Patterson, E. L., Holalu, S., Li, J., Johnson, G. A., Stanley, L. E., ... & Yuan, Y. W. (2018). Formation of periodic pigment spots by the reaction-diffusion mechanism. bioRxiv, 403600.
Jung, H. S., Francis-West, P. H., Widelitz, R. B., Jiang, T. X., Ting-Berreth, S., Tickle, C., ... & Chuong, C. M. (1998). Local inhibitory action of BMPs and their relationships with activators in feather formation: implications for periodic patterning. Developmental Biology, 196(1), 11-23.
Kondo, S., & Miura, T. (2010). Reaction-diffusion model as a framework for understanding biological pattern formation. Science, 329(5999), 1616-1620.
Medel, R., Botto-Mahan, C., & Kalin-Arroyo, M. (2003). Pollinator‐mediated selection on the nectar guide phenotype in the Andean monkey flower, Mimulus luteus. Ecology, 84(7), 1721-1732.
Sick, S., Reinker, S., Timmer, J., & Schlake, T. (2006). WNT and DKK determine hair follicle spacing through a reaction-diffusion mechanism. Science, 314(5804), 1447-1450.
Torii, K. U. (2012). Two-dimensional spatial patterning in developmental systems. Trends in Cell Biology, 22(8), 438-446.
Turing, A. M. (1990). The chemical basis of morphogenesis. Bulletin of Mathematical Biology, 52(1-2), 153-197.
Yamaguchi, M., Yoshimoto, E., & Kondo, S. (2007). Pattern regulation in the stripe of zebrafish suggests an underlying dynamic and autonomous mechanism. Proceedings of the National Academy of Sciences, 104(12), 4790-4793.
Yuan, Y. W., Sagawa, J. M., Frost, L., Vela, J. P., & Bradshaw Jr, H. D. (2014). Transcriptional control of floral anthocyanin pigmentation in monkeyflowers (Mimulus). New Phytologist, 204(4), 1013-1027.